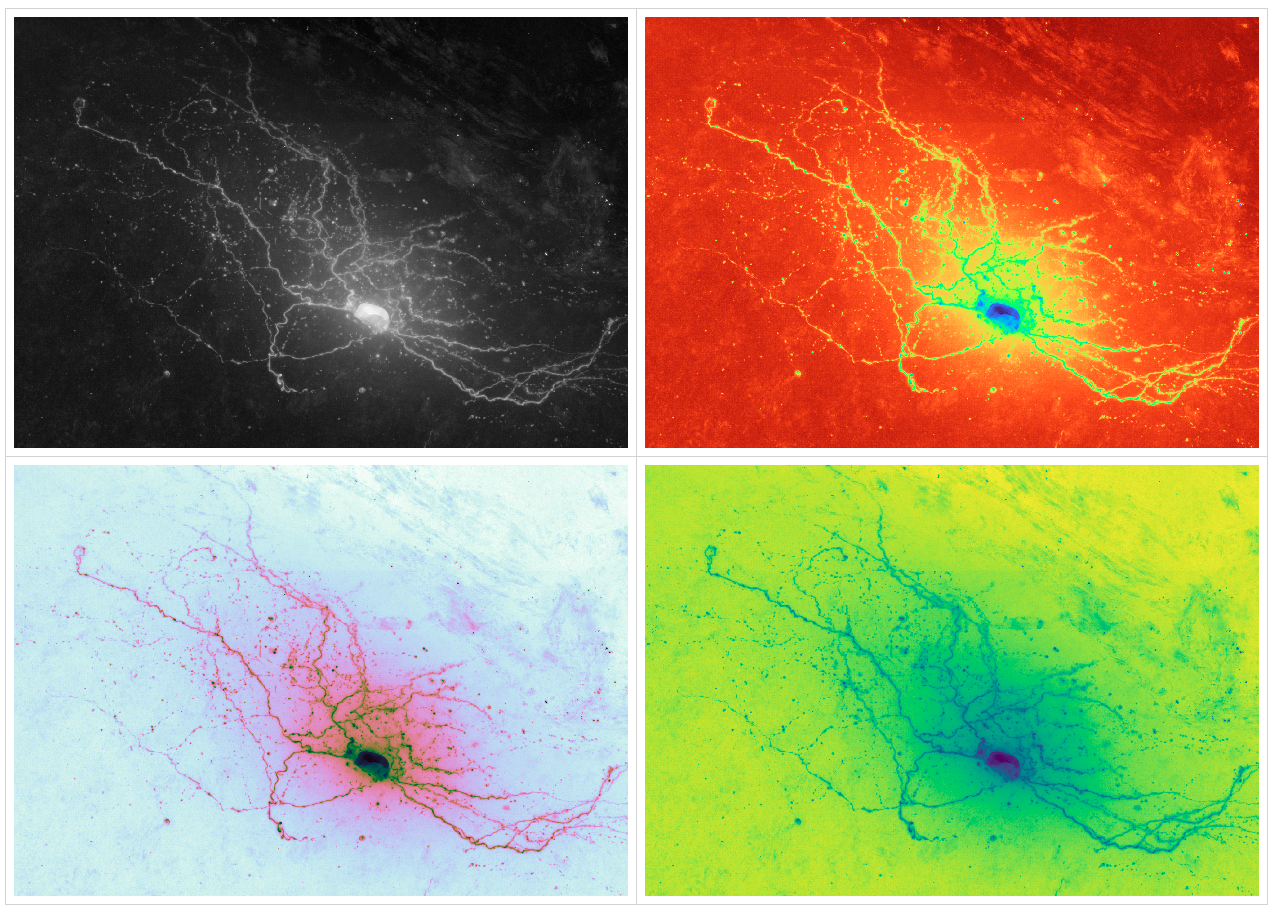
In neuroscience microscopy, "reconstruction" is a term of art for a digital model of a neuron which is derived from microscopy images.
The topic of this research project is the reconstruction of neurons imaged via brightfield microscopy, on Google's Colab. In the context of neuroscience, the brightfield modality is used to image a neuron internally stained with biocytin, rendering that single neuron floating opaque in a cleared volume of brain. The goal of this software project is to reconstruct the skeleton of the neuron via machine learning (DNNs, CNN, and recurrent variants there of).
This research is presented as an Apache 2.0 licensed repository of open source tools, brightfield_on_colab. The repository consists of Jupyter notebooks which run on Colab, Google's free Jupyter service such that any interested party can perform the same analysis as discussed herein.
Visualization
The analysis pipeline starts with raw images off of a microscope (either from a public repository or from personally uploaded-to-Colab image stacks). Various web-friendly visualization can be performed.
- Image stack navigation
- Static images
- Image pyramids (a la Google Maps)
- Animated GIF
- Stack dive as movie
- Minimum Intensity Projections (MinIP)
- Colorization
The visualizations also extend into research diagnostic tools for developing computer vision skeletonization algorithms for object recognition of the neurons in the brightfield image stacks.
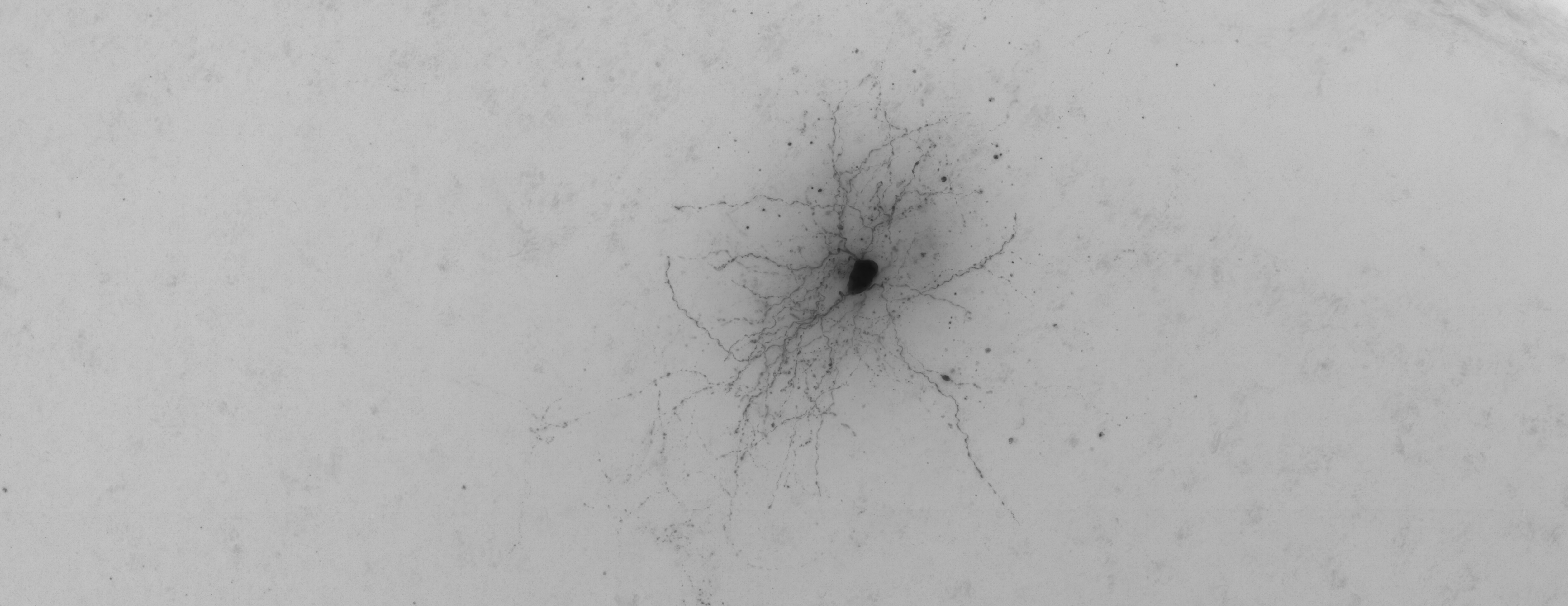
Skeletonization
"Skeletonization" is the process of taking the raw "grayscale" images of a
microscopy volume and constructing a stick figure skeleton
representation of a neuron's physical morphology. In terms of data
volume, this is a 3D to 1D reduction. The data structure of the
skeleton is semantic to the domain and as such is useful as input to
futher analysis. The skeletons (as *.swc files) represent the output
of the inital ETL
stage of microscopy pipelines.
Skeletonization is an open problem in computer vison and neuroscience. It is currently a bottleneck in neuroscience, necessitating long hours of manual image annotation. This would seem to be the sort of thing that recent developments in CNN would perform well at, yet none the less the extremely complex morphologies of neurons are proving difficult to completely recognize.
brightfield_on_colab provides diagnostic tools for researchers to evaluate the performance of skeletonization algorithms. Visually, there is a skeleton juxtaposer for comparing two skeletons, say a gold standard skeleton and another one produced via some experimental reconstruction algorithm. Volume unions provide more numerical scores.
Run on Colab
The main goal of this project is the development and distribution of software for producing skeletons (SWC files). The tools are open source, liberally licensed under Apache 2.0, impling there are no patents or other hidden restrictions. The software is intended to be industrial strength for use by industry.
The open source tools in brightfield_on_colab are presented as Jupyter notebooks that have been designed for deployment on Colab, where users perform analysis for free.
In this site, look for the following button to execute the code in the current page:
For example, click the Open in Colab button at the top of this page and then Run the notebook to get "Hello, World!"
print("Hello, World!")
Implementing the algorithms to work on Colab addresses the distribution problem, as well as the provisioning of compute resources to execute the code in the notebooks.
In this way, Colab can be used as a lightweight form of reproduceable research: analysis can be worked out on Colab and published on bioarxiv.org, where the PDF could link back to a Jupyter Notebook running on Colab.
Project status
See appendix: Project status
