Review articles from the previous decade [Marx 2012, Blackman 2014, Parekh 2014] are available as primers.
Neuronal Morphology goes Digital: A Research Hub for Cellular and System Neuroscience (2013):
Biocytin and Biotinamide (Neurobiotin)
Biocytin is a small conjugate of biotin and lysine naturally found in eukaryotic organisms. Due to its low molecular weight and biocompatibility, it constitutes a valuable tool in whole-cell or juxtacellular recordings, as it is incorporated into living neurons without perturbing ionic balance or membrane properties. Biocytin stains both axons and dendrites and is not transported trans-neuronally... Both biocytin and biotinamide have high affinity for avidin, and the tissue is processed post-fixation using an avidin-biotin-peroxidase complex followed by DAB reaction...Bright field microscopy
The majority of dendritic and axonal morphologyreconstructions to date are based on bright field microscopy (Halavi et al., 2012), due to its broad compatibility with histological staining methods. In conventional bright field microscopy, as the name suggests, the tissue background is illuminated by transmitted light, whereas the stained neuron absorbs the light and is visible in dark contrast against the bright background. However, for certain applications or depending on user preference, simple image processing can be employed to invert this contrast (Myatt et al., 2012). Thus this modality should be more precisely referred to as trans-illumination or transmitted light microscopy. Unlike confocal microscopy, which requires fluorescent labels, bright field microscopy can visualize Golgi stain preparations and intracellular labels like biocytin. Even neurons labeled with fluorescent markers can be permanently labeled by DAB reaction and imaged with bright field microscopy. Moreover, the ability to enhance the signal intensity by counterstaining renders bright field microscopy largely unsurpassed for reconstructions of whole axonal arbors up to the very thin (and typically faint) terminals.
Brightfield is the simpliest of microscopy modality. Simple and old also means mature and cheap. Coincidentially, brightfield plus biocytin staining is a high quality neuron imaging technique. For these and other reasons brightfield is the most widely adopted.
None-the-less, brightfield data is dirty conpared to, say, electron microscopy (EM) data and as such makes it more difficult to apply modern computer vision techniques such as CNNs, U-Net, Flood-Filling Networks (FFNs), etc. That is the topic of this project: how to backport any applicable innovations from other microscopy modalities (e.g. EM or flourescent). How to pre-process brightfield data such that it can be fed to existing software from, say, EM.
After surveying any applicable prior art, innovations specific to brightfield are on the cutting edge of the art. As such SOTA will improve over time. This needs to be measured via evalution datasets. All the code in this project has been designed to run on Colab. As such, Colab is used as the standardized execution platform for performing evalutions.
Nature of the data
Brighfield modality data are noisy compared to, say, electron microscopy (EM). For example, in the following images only a small region of the image stack are clear. In the following example depths 72 and 109 are where the images are clearest. Each stack has a different region of focus depending on how the specimen is mounted on the microscope's stage. The stage moves; the camera does not.
Pull some sample from throughout a random z-stack at various depths. The bluriness increases smoothly throughout the stack as the foreground object (a biocytin-stained neuron) moves through the scopes field of view. This is an artifact of the stained specimen on the glass slide moving throught the microscope's stationary field of focus.
In brightfield microscopy, to get a 3D stack of images the specimen is not serially sectioned (think: deli meat slicer, which is not the case with brighfield). Rather the intack slab of brain is passed through the optical field of focus of the microscope. The stage that the glass slice is mounted on is moved along the Z-axis of the microscope. The 3D "scanning" is a series of photos as the specimen is moved.
While moving along the Z-axis eventually the stained specimen is too far from the focal region. It looks blurry. This effect is seen at both end of the image stacks in the brightfield data.
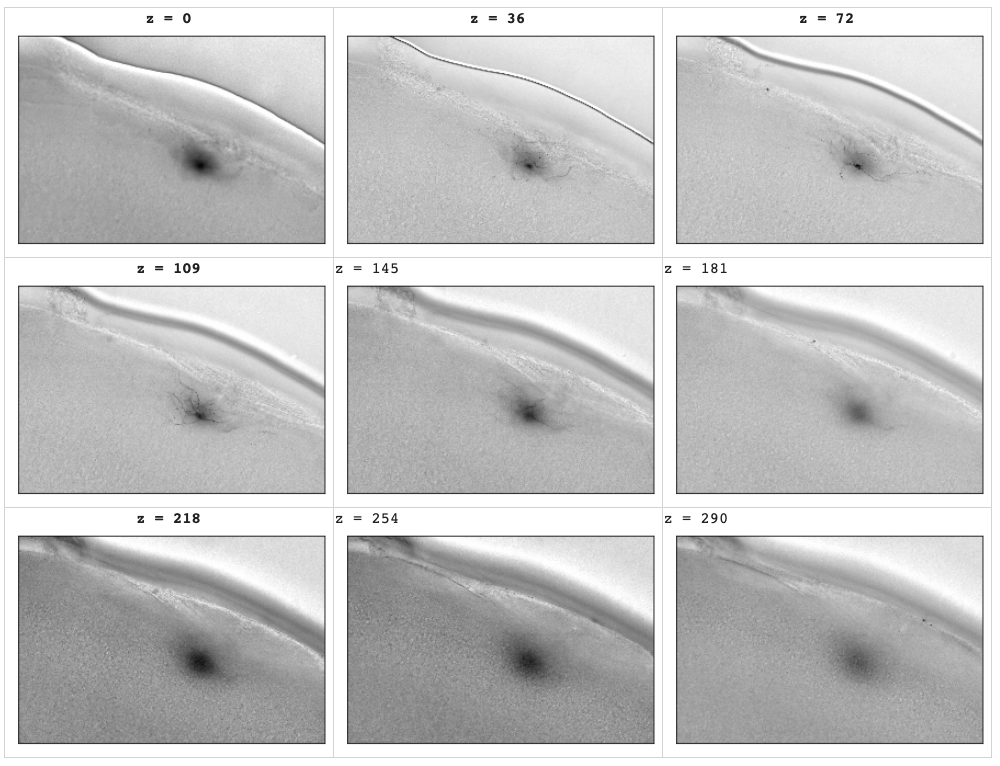
A silly analogy would be to think of a biocytin stained neuron as if it were a tourist atop an open-topped bus (the microscope stage). A security CCD (the microscope's camers) takes a series of photos of the neuron as it it driven though the camera's field of view. Much of the footage is very blurry.
References
Example recent historical documents on the state of the field.
- 2012: Improved biocytin labeling and neuronal 3D reconstruction in Nature Protocol 7(2):394-407
- 2014: A comparison of manual neuronal reconstruction from biocytin histology or 2-photon imaging: morphometry and computer modeling
- Front. Neuroanat., 11 July 2014 | https://doi.org/10.3389/fnana.2014.00065
