In connectomics, Google has two very interesting 2017 codebases, Flood-Filling Networks (FFNs) and Neuroglancer. The former is a neural network which performs image segmentation of volumes of brain slices to construct stick figure skeletons and 3D mesh models of detected neurons; the latter is an in-browser visualizer of the former’s results. Google’s Neuroglancer demo (live in-browser) works in any modern release of Chrome or Firefox.
In Neuroglancer, after about a minute of basic navigation – clicking and dragging on colored blobs – a user quickly gets the idea: Neuroglancer is a 3D spatial navigator of 2D black and white brain slice images, with “augmented reality” 3D mesh models of FFN-detected neurons, each highlighted with a distinct color.
In November 2017, in partnership with the Max Planck Institute, Google released the Neuroglancer source code to support their paper, High-precision automated reconstruction of neurons with flood-filling networks, which was eventually published in Nature Methods. (Earliest related paper yet found is from November 2016.)
The crux of the Flood Filling Networks (FFNs) work is a network architecture – essentially a CNN enhanced with a RNN – that can do colored image segmentation of black-and-white (electron microscopy) brain images, thereby reconstructing the imaged neurons in 3D. These FFNs (and at least two other examples, discussed herein) are ML-based automation tools with the potential to save many hours of tedius manual neurite tracing.
Along with the FFN algorithm, Google AI simultaneously released Neuroglancer, which is in-browser software for viewing the output of a FFN. Both codebases (neuron detector algorithm and visualizer) are licensed under Apache 2.0. For more detail see Google’s July 2018 introductory blog post, Improving Connectomics by an Order of Magnitude.
Status
Code (Apache-2.0 licensed) code was taken from Google's repro: ffn_inference_demo.ipynb and used here to seed probing if it could be run on Colab.
This issue is being tracked on GitHub as #73.
Intro papers
The first paper is the 2016 pre-print, Flood-Filling Networks (on arxiv).
The main FFN paper is entitled, "High-precision automated reconstruction of neurons with flood-filling networks" but the place to start reading might be Google's announcement blog post, Improving Connectomics by an Order of Magnitude.
- pre-print on biorxiv
- Nature article ($)
- Nat Methods. 2018 Aug;15(8):605-610.
doi: 10.1038/s41592-018-0049-4.
2018 Jul 16.
- Nat Methods. 2018 Aug;15(8):605-610.
Algorithm illustration
The above illustrates a run of FFN which (flood) fills one neuron at a time. The red is what is segmented/filled. The yellow is the inference algorithm working within a small cuboid of active field-of-view.
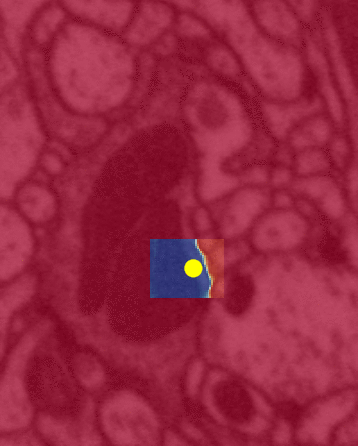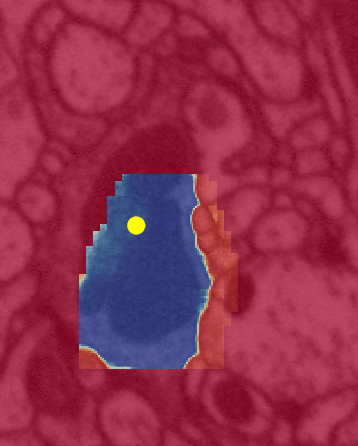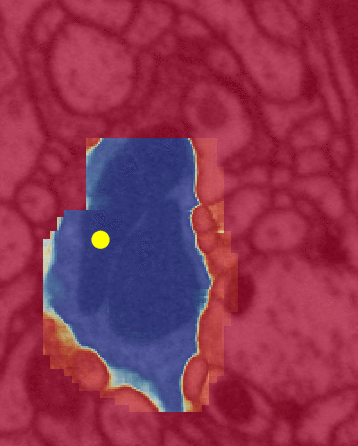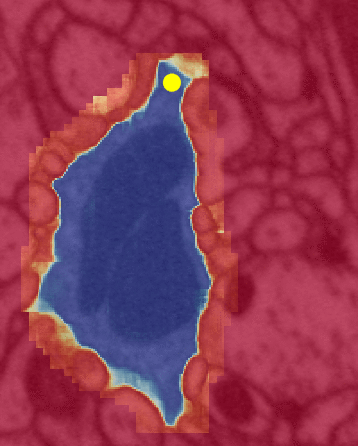
Left is a close up view of the same algorithm. The color yellow is the same in both (inference attention). Unfortunately, between the two illustrations black => red and red => blue.
The yellow dot is the FFN’s attention mapping (flood filling) within a single neuron by bouncing off the cell’s walls.
The following image sequence illustrates the sequential nature of the algorithm (in a dense volumetric EM context), i.e. cells are segmented one-by-one randomly.
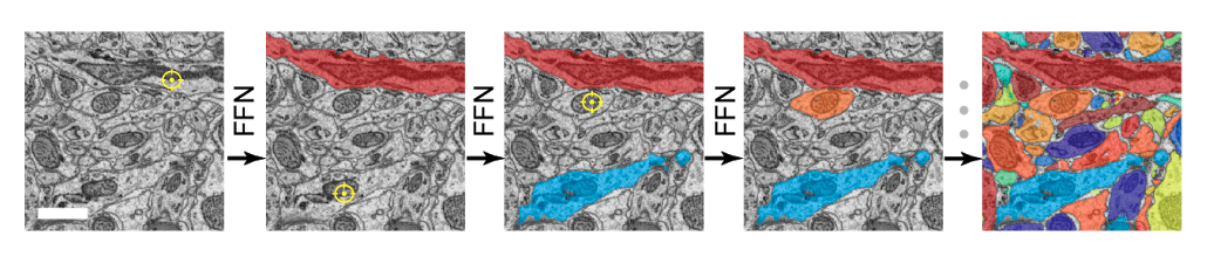
FFNs have been successfully applied to dense volumetric EM data by Google and Max Panck. The question is: can this technology be adopted to single-neuron brightfield image stacks.
Labeling training data
VAE output to train FFN supposedly rather easy.
A VAE architecture:
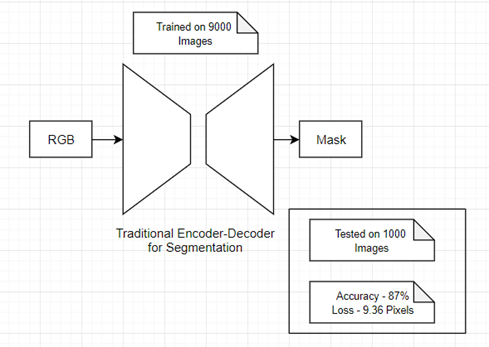
Used to bootstrap training data for FNN.

That would seemingly reduce training costs: "architecture is computationaly easy to train."
FFN adoptions
Learning cellular morphology with neural networks, Nature Communications volume 10, Article number: 2736 (2019).
Say you wanted to remove all the glia from a chuck of dense volumetric EM imaged brain, making it easier to see certain neurons (glia are 90% of the cells in brains). Sure would be handy to have a CNN-ish classifier tool to filter out all the red glias.
References
Code
- Main repo: https://github.com/google/ffn
- (Apache-2.0)
Papers
First paper: FFNs
Pre-prints:
Blog posts and such